The main interaction partners of the ribosome
To carry out translation of an RNA template into a protein, the ribosome is dependent on a large set of molecules – mRNAs, tRNAs, protein factors and energy carriers.
Messenger RNA (mRNA) contains the protein sequence
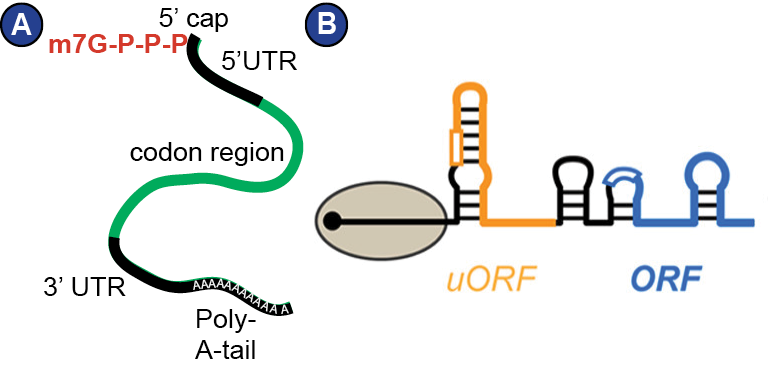
The information on the sequence of amino acids is encoded in the DNA of a cell and has to be transferred to RNA first (paradigm of molecular biology (Crick, 1970)). Such RNA, which serves as template for protein synthesis, is called messenger RNA (mRNA). Its production involves transcription by polymerase II and processing steps such as splicing, polyadenylation and capping (Meister, 2011). The mRNA pool accessible to the ribosome is strictly controlled by a balance between production and decay, variation of polyadenylation, storage in P-bodies, and many other mechanisms that are not well understood yet (Meister, 2011). In eukaryotes, mature mRNA is characterized by a 5’ cap, a 5’ UTR (untranslated region), the coding region, a 3’ UTR, and a poly-A-tail ( Figure 4 ). Notably, the mRNA is intensely decorated with proteins that regulate its transport, stability, decay and translation, for example poly-A-binding proteins (PABPs). Thus, mRNA really is a messenger ribonucleoprotein (mRNP) (Mitchell and Parker, 2014). Moreover, mRNA forms secondary structure that influences its interaction with the ribosome ( Figure 4 B). The entirety of the cellular mRNAs produced for translation at a given time-point is called transcriptome, and the actually translated mRNAs in a cell at a given timepoint constitute the translatome.
Transfer RNAs (tRNAs) allow for exact selection and incorporation of amino acids
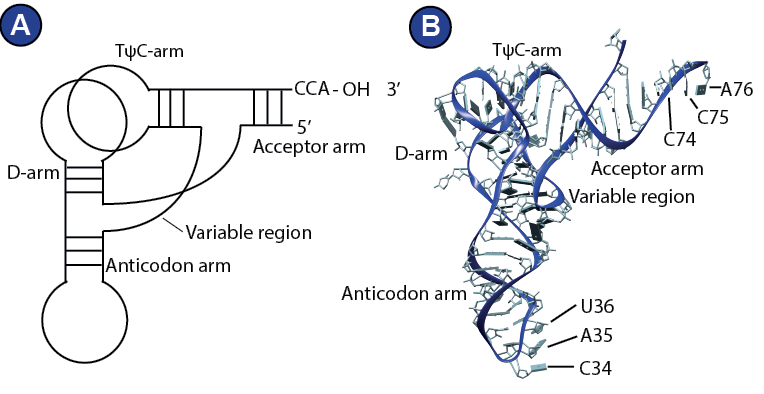
tRNAs are specialized RNA molecules that usually consist of around 76 nucleotides. Their role in translation is to deliver the amino acids to the ribosome. Structurally, a tRNA consists of two parts, top and bottom, which organize into an L-shaped tertiary structure (Gesteland, R.F., Cech, T.R., 1999).
The top half comprises the TΨC-arm and the acceptor arm with the CCA-end, which is loaded with the amino acid by specialized enzymes, aminoacyl-transferases. There are elongator tRNAs and initiator tRNAs. Initiator tRNAs (tRNAi) are always loaded with a methionine. The bottom half of a tRNA consists of the D-arm and the anticodon loop, which can decode the mRNA ( Figure 5 ). The tRNA nucleotides are often and extensively modified and these modifications are thought to be important for its structure and function (Meister, 2011). The life of tRNA outside of the ribosome is object of intensive research and much less is known about it than about its role on the ribosome. The modification of a tRNA, for example, is complex and includes big protein complexes (Dauden et al., 2017). Further, like the mRNA, the tRNA does not exist in the cytosol as free and naked RNA molecule. The eukaryotic cell operates an elaborate system that channels the tRNAs to and from the ribosome, involving aminoacyl-transferases and probably more factors (Andersen et al., 2006; Mirande, 2010; Stapulionis and Deutscher, 1995).
Translational GTPases tune the energy landscape of translation
Many protein factors are involved in translation by direct interaction with the ribosome, the mRNA or the tRNA. A special subgroup of protein factors involved in translation are translational GTPases. They are GTP-hydrolyzing proteins that control key steps of translation; for example the decoding-specific eukaryotic elongation factor 1A (eEF1A), or eukaryotic elongation facter 2 (eEF2), which is necessary for efficient translocation, eukaryotic release factor 3 (eRF3), which is responsible for stop-codon recognition and termination, and eukaryotic initiation factor 5B (eIF5B), which mediates subunit joining (see section ‘translation cycle’).
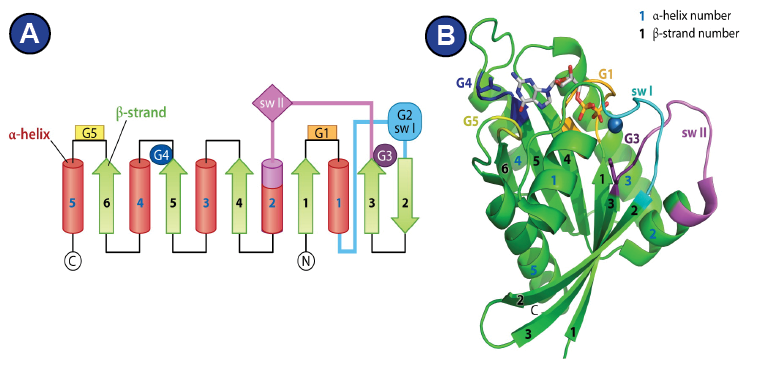
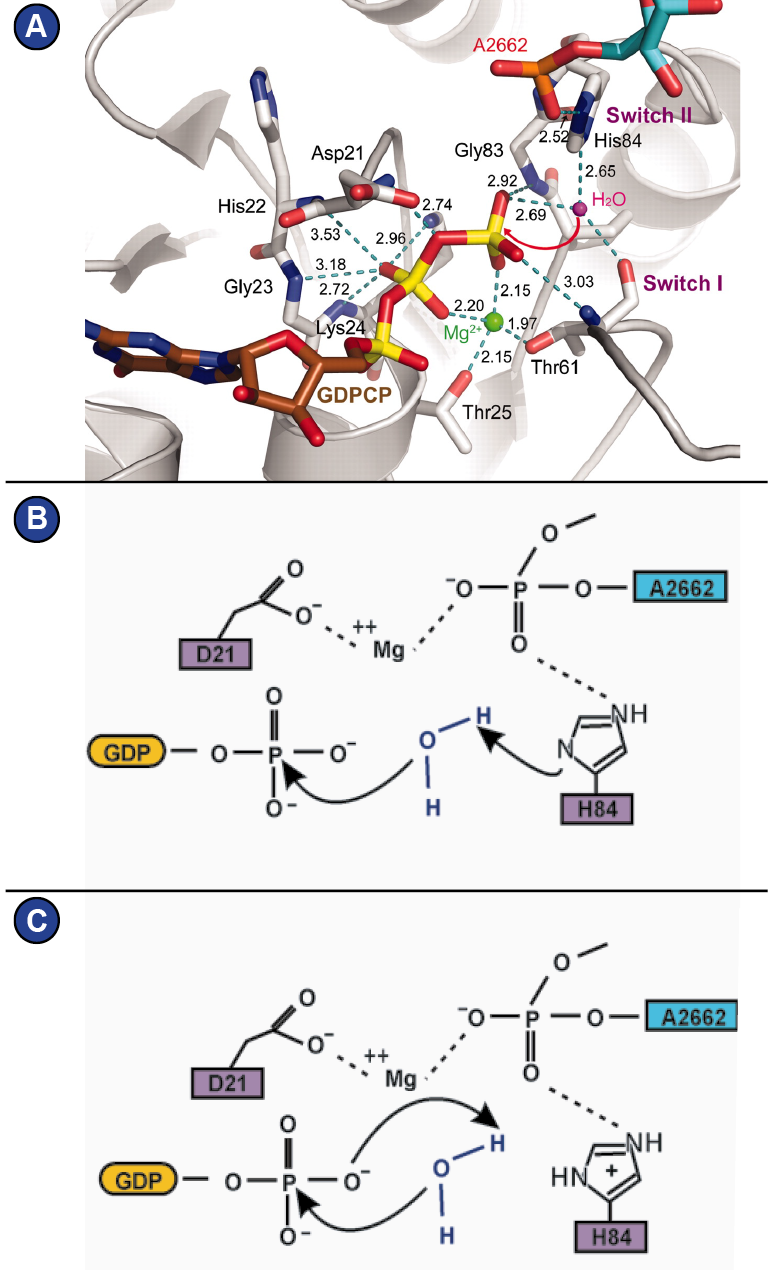
Structurally, they all have in common the GTP-hydrolyzing domain (G-domain), which forms an alpha-beta-propeller and contains the nucleotide binding pocket (Wittinghofer and Vetter, 2011). Five characteristic G-motifs (G1-G5) and three loops (P-loop, switch I, switch II) organize around the bound nucleotide and characterize the G-domain ( Figure 6 ; Supplemental Figure 6 shows alignment of the G-domain of EF-G/eEF2 from different species). The G1 motif is also known as Walker A motif and is located within the P-loop. It is responsible for phosphate binding. The Mg2+ that is required for nucleotide binding is positioned by the G2 motif and the G3 motif. The G2 motif is part of switch I, whereas the G3 motif (also known as Walker B motif), is located within switch II (Bourne et al., 1991; Wittinghofer and Vetter, 2011) (Figure 6). GTP-hydrolysis is achieved by a nucleophilic attack of an activated water molecule and is assisted by the SRL of the ribosome, which serves as GTPase-activating factor for the translational GTPases. Depending on how one interprets the role of a histidine of switch II (His 84 in eF-Tu, His 108 in eEF2) ( Figure 7 A), two mechanism are being proposed that lead there: 1) A general base mechanism, in which the histidine abstracts a proton from the water molecule ( Figure 7 B), and 2) a substrate-assisted mechanism, in which GTP itself abstracts a proton from the water molecule while the histidine serves as allosteric enhancer of this process (Figure 7C) (Liljas et al., 2011; Maracci and Rodnina, 2016; Schweins et al., 1995; Voorhees et al., 2010).